Kostrouch Lab
Contact: zdenek.kostrouch@lf1.cuni.cz
Lab Members
Zdeněk Kostrouch, Marta Kostrouchová, Petr Yilma
Ahmed Chughtai, Jan Novotný, Filip Kaššák, Kateřina Šušlíková, Tomer Rosenberg.
Laboratory infrastructure and methods
Laboratory for molecular and cell biology, genetics and genomics of invertebrate model organisms Caenorhabditis elegans, Trichoplax adhaerens, complexgenomics and genetics, genome editing including de novo gene design. Genome editing in selected plant systems (presently Cannabis sativa).
Specialized methods are performed in cooperation with scientists from Research Core Facilities of BIOCEV https://www.biocev.eu/en/infrastructures-and-core-facilities and currently include advanced photonics (Laser scanning microscopy, Fluorescence-lifetime imaging microscopy, Coherent Anti-Stokes Raman Scattering Microscopy) and advanced proteomics (Mass Spectrometry).
Images from our current research:
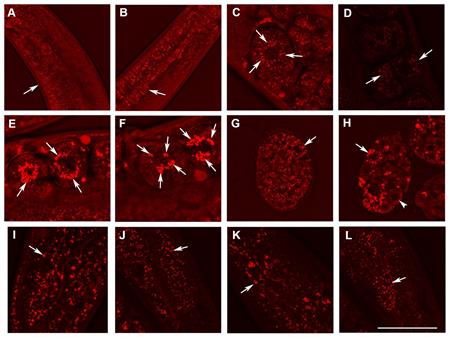
Picture 1) Detection of fat containing structures in live C. elegans with edited Perilipin gene (plin-1) and in controls. Analysis of lipid containing structures in live wild type animals and in animals with disrupted W01A8.1 by CARS microscopy. CARS microscopy was kindly performed in cooperation with Dr. Zhongxiang Jiang and Leica Microsystems CMS GmbH (Mannheim, Germany). Panels E and F show enlarged fat containing compartments that likely visualize a new mechanism of regulated transport of lipids and other associated molecules to daughter cells during cell division. For details, please see https://peerj.com/articles/1213/

Picture 2) Sea water aquarium containing cultures of Trichoplax adhaerens.
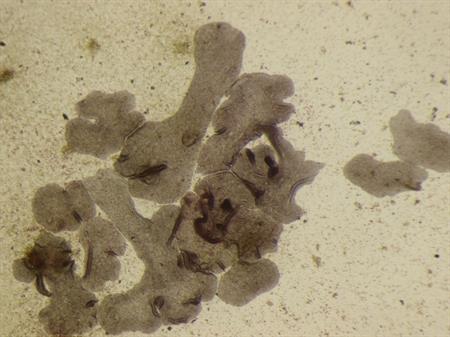
Picture 3) A group of socializing Trichoplax adhaerens feeding on Porphyridium cruentum.
Trichoplax adhaerens represent an organism that is likely to exist at the base of Metazoan evolution. The regulation of gene expression by the orthologue of RXR is conserved in this basal species. For details, please see https://peerj.com/articles/3789/.
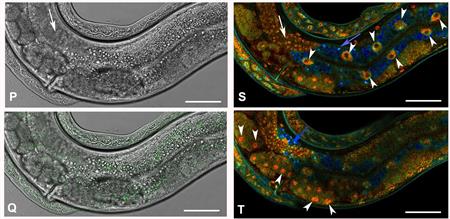
Picture 4) Detection of edited MDT-28 in Caenorhabditis elegans using Fluorescence-lifetime imaging microscopy. For details, please see https://peerj.com/articles/3390/.
Lab scope and current directions
1. Targeting specific gene expression for development of new therapeutic strategies
Regulation of gene expression in invertebrate model organisms (Caenorhabditis elegans, Trichoplax adhaerens) and in human cell cultures and tissues.
2. Bioengineering – Genome editing for betterment of life
Genome editing of plant tissue and cell cultures for bioengineering of new plants and plant cell systems with the potential of production of new molecules with specific features (binding of specific molecular structures for diagnostics and therapies and for establishment of new plant based food sources).
Research concepts:
1. Regulatory connection between cell structural states and gene expression: structural proteins and their interactors modulate regulatory signals of transcription factors and adapt gene expression to specific cell structural state.
2. Bioengineered plants and plant cell cultures can provide human food and molecules with specific properties better than systems exploiting animals.
Research statements:
Biology and medicine are changing very fast during the recent years. Nowadays, we would give our laboratory a new name: Laboratory of Imaginative Biology. In the past, we could use only what Nature offered us. We could amplify genes or separate compounds, and search for their effects in natural or modified versions in experimental systems.
Nowadays, we can do almost anything that we can think of, everything that we can imagine. We can create new genes, introduce them to living systems, even to adult fully grown or old organisms. We can introduce such artificial genes to almost any organism (that we are able to cultivate in the laboratory) to almost any chosen place in the genome and design their manageable regulations. In a sense, even this is derived from what Nature offers but the extent of human intervention is greater, more powerful and for science advancing faster.
Unfortunately, as everything in the human world, these imaginations can be good or bad, ethical or unethical. We aim and do our best to materialize imaginations that are ethical in the most absolute dimensions: helping to decrease or eliminate animal suffering and leading to betterment of life.
We use invertebrate organisms for analysis of regulatory pathways and plant systems for production of compounds that used to be produced in animals.
We work on the concept of the free proteome code, a concept that assumes the regulatory roles of proteins that cannot be assembled in cell structures and act as regulators of gene expression. We also worked on ancient systems of gene expression regulation by nuclear receptors and came up with the concept of food constituents as the original endocrinologically active substances that still influence our lives but are obscured by more powerful modern regulations.
Our ongoing projects include the role of mediator subunits in regulation of cell stemness in normal and cancerous cells, production of specifically designed plant and bacterial nanobodies with specific binding properties and plant based fully balanced food for people and their animal companions.We cooperate with Biocev research groups and our friends from around the World on these research programs.
Selected Publications
Kostrouchová M, Kostrouchová V, Yilma P, Benda A, Mandys V, Kostrouchová M. Valproic Acid Decreases the Nuclear Localization of MDT-28, the Nematode Orthologue of MED28. Folia Biol (Praha). 2018;64(1):1-9.
Novotný JP, Chughtai AA, Kostrouchová M, Kostrouchová V, Kostrouch D, Kaššák F, Kaňa R, Schierwater B, Kostrouchová M, Kostrouch Z. Trichoplax adhaerens reveals a network of nuclear receptors sensitive to 9-cis-retinoic acid at the base of metazoan evolution. PeerJ. 2017 Sep 29;5:e3789. doi: 10.7717/peerj.3789. eCollection 2017.
Kostrouchová M, Kostrouch D, Chughtai AA, Kaššák F, Novotný JP, Kostrouchová V, Benda A, Krause MW, Saudek V, Kostrouchová M, Kostrouch Z. The nematode homologue of Mediator complex subunit 28, F28F8.5, is a critical regulator of C. elegans development. PeerJ. 2017 Jun 6;5:e3390. doi: 10.7717/peerj.3390. eCollection 2017.
Kaššák F, Hána V, Saudek V, Kostrouchová M. Novel Mutation (T273R) in Thyroid Hormone Receptor β Gene Provides Further Insight into Cryptic Negative Regulation by Thyroid Hormone. Folia Biol (Praha). 2017;63(2):60-66.
Chughtai AA, Kaššák F, Kostrouchová M, Novotný JP, Krause MW, Saudek V, Kostrouch Z, Kostrouchová M. Perilipin-related protein regulates lipid metabolism in C. elegans. PeerJ. 2015 Sep 1;3:e1213. doi: 10.7717/peerj.1213. eCollection 2015.
Zima V, Šebková K, Šimečková K, Dvořák T, Saudek V, Kostrouchová M. Prorenin Receptor Homologue VHA-20 is Critical for Intestinal pH Regulation, Ion and Water Management and Larval Development in C. elegans. Folia Biol (Praha). 2015;61(5):168-77.
Kostrouch D, Kostrouchová M, Yilma P, Chughtai AA, Novotný JP, Novák P, Kostrouchová V, Kostrouchová M, Kostrouch Z. SKIP and BIR-1/Survivin have potential to integrate proteome status with gene expression. J Proteomics. 2014 Oct 14;110:93-106. doi: 10.1016/j.jprot.2014.07.023. Epub 2014 Aug 1.