Janoštiak Lab
Group name: Laboratory of Molecular Oncology
Group Leader: RNDr. Radoslav Janoštiak, Ph.D.
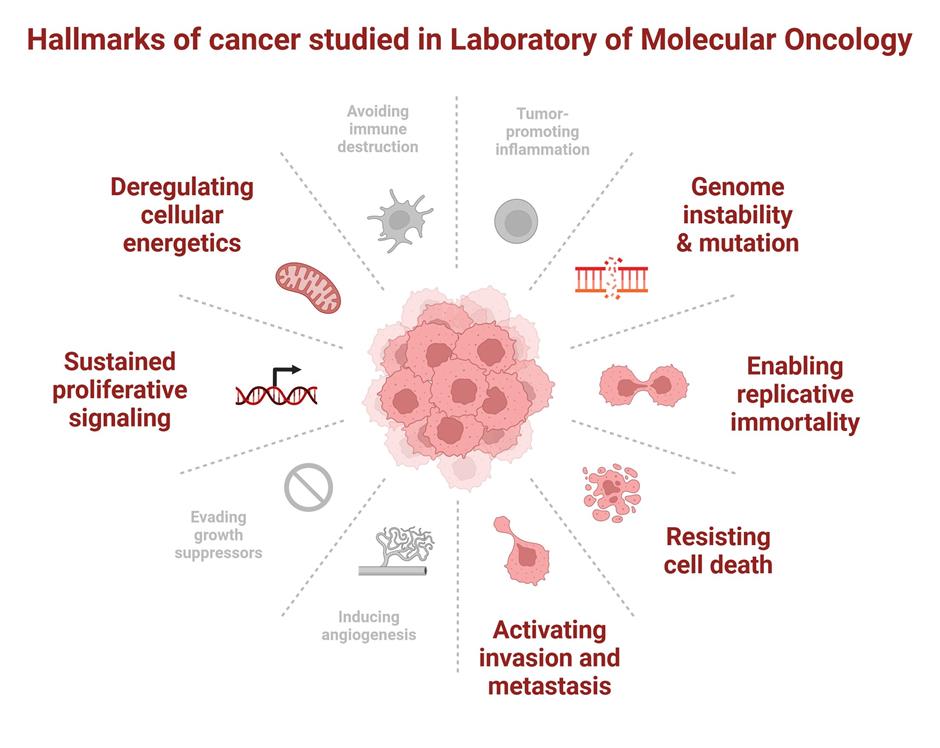
Cancer is a multifactorial disease with multiple driving mechanisms, following the Darwinian survival of the fittest theorem. Despite significant inter- and intra-tumoral heterogeneity, there are common hallmarks shared across the board including sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, activating invasion and metastasis, reprogramming energy metabolism, evading immune destruction, promoting tumor-associated inflammation, and deregulating cellular energetics.
Research conducted in the Laboratory of Molecular Oncology focuses on several of these hallmarks in the context of the following areas:
Regulation of cell cycle arrest maintenance and exitThe ability to dynamically control the cell proliferation is crucial for homeostasis maintenance in multicellular organisms. There are three main cellular states, proliferative, quiescent and senescent, and each cell has to integrate different environmental cues in order to keep the homeostasis. Disruption of this balance leads to pathological conditions such as cancer. However, even cancer cells have to be able to regulate the transition between proliferation and quiescence in order stimulate the metastatic spread or to survive environmental stress (such as low nutrients or therapy). Our laboratory is studying this transition at the level of proteomics and posttranslational modifcations on the model of triple negative breast cancer.
Protein homeostasisRegulation of stability and turnover of the proteins is a key feature of the living cells, and it responds dynamically to various extracellular and intracellular signals. At the global level, proteosynthesis responds to nutrient availability and environmental stresses. On the other hand, proteins are selectively degraded through ubiquitin proteasome degradation system. In our laboratory, we study the regulation of cap-dependent and cap-independent translation in cancer, as well as ubiquitin-mediated degradation of regulators of cell cycle and survival.
Cancer cell metabolism: A common feature of cancer cells is the ability to alter their metabolism to maintain the production of ATP and macromolecules required for cell growth, division, and survival. Besides well described Warburg effect, cancer cells adapt also other metabolic pathways, among them metabolism of fatty acids. In our laboratory we are studying the regulation of fatty acid metabolism in respect to energy and metabolic intermediates production.
Additionally, we are working on identification of diagnostics and prognostic biomarkers for pancreatic cancer and endometriosis.
Methodologically, we utilize wide range of techniques including but not limited to DNA manipulation (cloning, site directed mutagenesis, RNAi, CRISPR), general and advanced MS-based proteomics, RNA sequencing, 3D organoid culture and analysis of patient-derived tissue samples and cells.
Group profile at BIOCEV website HERE.